Yeast mutation. Saccharomyces Genome Database 2022-12-16
Yeast mutation
Rating:
6,5/10
1643
reviews
Yeast mutation is the process by which changes occur in the genetic material of yeast cells. These changes can be either beneficial or harmful to the yeast, and they can occur spontaneously or be induced artificially. In either case, the mutations can have significant impacts on the characteristics and behavior of the yeast cells, which can be of great importance in a variety of different contexts.
One of the key ways in which yeast mutation can be important is in the production of new yeast strains with desired characteristics. For example, by inducing mutations in yeast cells and selecting for cells with specific characteristics, it is possible to create yeast strains that are more efficient at producing ethanol or other products, or that are more resistant to certain types of stresses. This process of mutagenesis and selection has been used extensively in the development of new yeast strains for use in the food and beverage industry, as well as in other applications.
Another important context in which yeast mutation can be significant is in the study of genetics and molecular biology. By inducing mutations in yeast cells and studying the effects of these mutations on the cells' behavior and characteristics, scientists can learn more about the role of specific genes and gene products in various cellular processes. This type of research has led to many important insights into the underlying mechanisms of yeast cell biology and has helped to inform our understanding of these processes in other organisms as well.
Finally, yeast mutation can also be important in the study of evolution. By studying the changes that occur in yeast cells over time, scientists can learn more about how evolution works and how different organisms adapt to changing environments. This type of research can provide valuable insights into the processes that drive the evolution of all living things, including humans.
In summary, yeast mutation is a complex and multifaceted process that can have significant impacts on the characteristics and behavior of yeast cells. By understanding more about yeast mutation and its effects, we can better understand the underlying mechanisms of genetics and molecular biology, and we can use this knowledge to create new yeast strains with desirable characteristics and to study the process of evolution itself.
petite mutation

The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. It has inherited normal mitochondrial DNA from wild-type parent, which is replicated in the offspring. MAT locus is similar to that in S. New vectors for simple and streamlined CRISPR—Cas9 genome editing in Saccharomyces cerevisiae. Asexual experimental evolution of yeast does not curtail transposable elements. When an a cell cuts the MAT a allele present at the MAT locus, the cut at MAT will almost always be repaired by copying the information present at HML.
Next
Studying the Ballistics of Yeast Mutagenesis

Codon-by-codon modulation of translational speed and accuracy via mRNA folding. A mutant is purged if its fitness is lower than a preset cutoff such as 0. The lower and upper edges of a box represent the first qu 1 and third qu 3 quartiles, respectively, the horizontal line inside the box indicates the median md , the whiskers extend to the most extreme values inside inner fences, md ± 1. The Neutral Theory of Molecular Evolution Cambridge Univ. Its patterns of mutation, however, turned out to be unique. The DEAD-Box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality.
Next
Mating of yeast
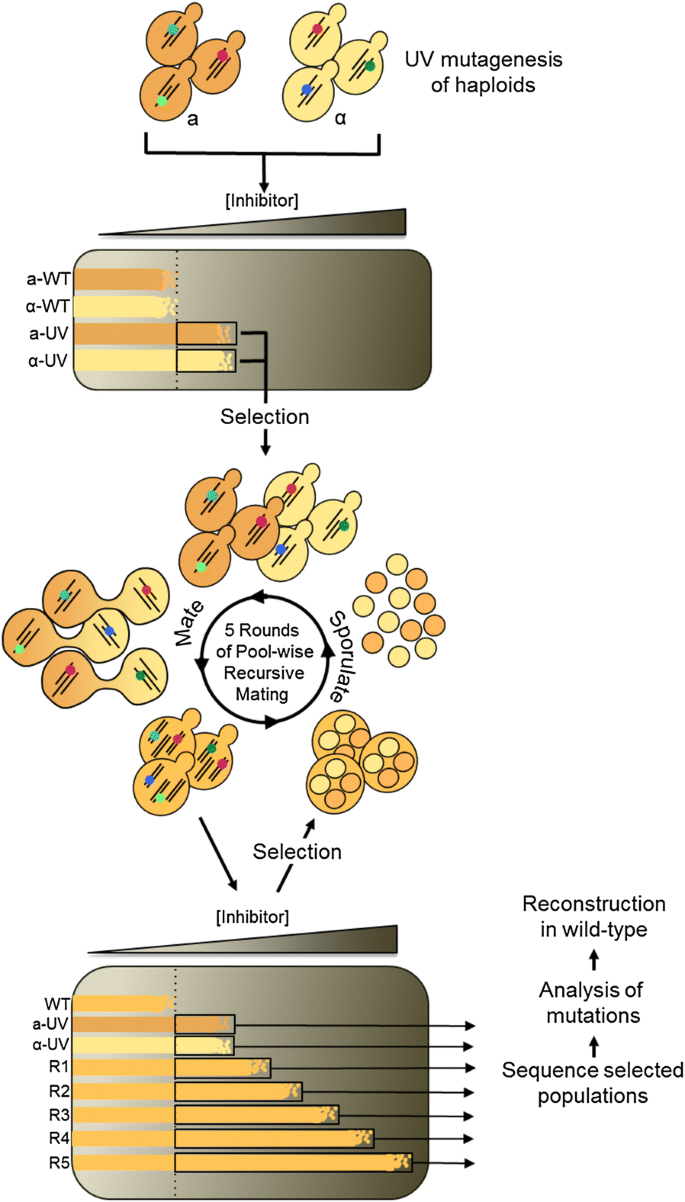
So that they could look at the full spectrum of mutations arising in the mutator strains, Stirling and coworkers decided to use whole genome sequencing instead to detect them. The nonsense-mediated decay RNA surveillance pathway. The different gene expression patterns of haploids and diploids are again due to the MAT locus. Science 342, 1367—1372 2013. However, not only does the mating decision need to be conservative in order to avoid wasting energy , but it must also be fast to avoid losing the potential mate.
Next
Revealing mutations in yeast

Comparing the mutation rates and spectrums of these two model organisms informs researchers' assumptions about mutation relevant to human health. The rMFS of a mutant is its mRNA folding strength i. Science 324, 255—258 2009. P-values are from two-tailed t-tests. Cell 160, 1111—1124 2015. Diploid cells result from the mating of an a cell and an α cell, and thus possess 32 chromosomes in 16 pairs , including one chromosome bearing the MAT a allele and another chromosome bearing the MATα allele. Significant impact of protein dispensability on the instantaneous rate of protein evolution.
Next
Synonymous mutations in representative yeast genes are mostly strongly non

USA 113, E6117—E6125 2016. Haploid cells only contain one copy of each of the 16 MAT either MAT a or MATα , which determines their mating type. However, while all of the mutator strains had accumulated mutations, the different types of mutation were in different proportions. This results in MAT being repaired to the MATα allele, switching the mating type of the cell from a to α. Mutator strains tend to look similar from the outside; many are deficient in DNA replication and repair pathways.
Next
Saccharomyces Genome Database

USA 115, E4940—E4949 2018. YEASTRACT+: a portal for cross-species comparative genomics of transcription regulation in yeasts. HML and HMR loci and the active MAT locus on yeast chromosome III. The frequency at which S. The vast majority of environmental and clinical isolates of C. Thus it appears that, in nature, mating is most often between closely related yeast cells.
Next
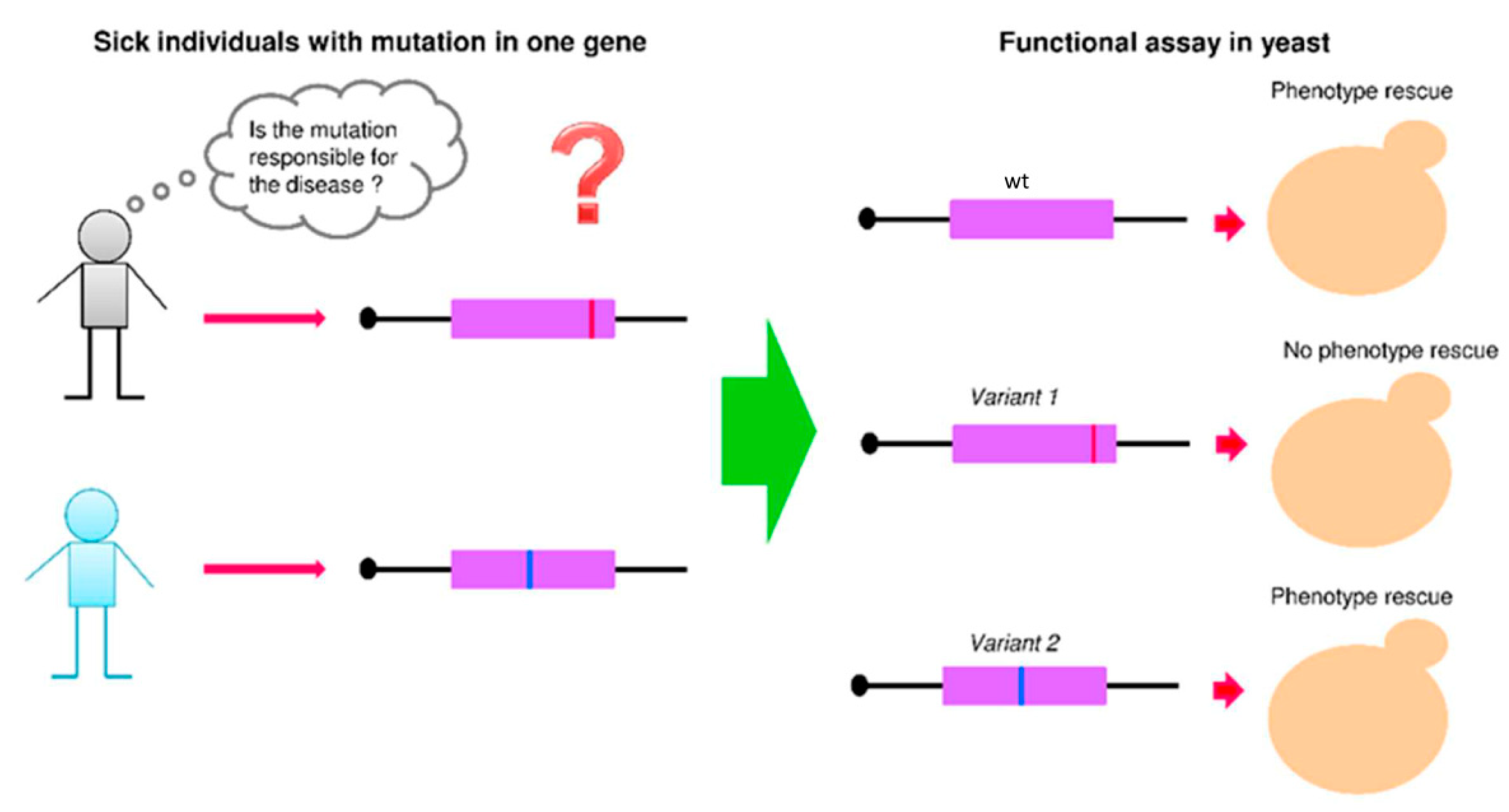
Segregational petites pet— : mutants are created by nuclear mutations and exhibit Mendelian 1:1 segregation. Biochimica et Biophysica Acta BBA - Bioenergetics. The MAT locus is usually divided into five regions W, X, Y, Z1, and Z2 based on the sequences shared among the two mating types. Transcriptome-wide analysis of roles for tRNA modifications in translational regulation. Cell 175, 544—557 2018. Given that some of these same genes have been found to be mutated in cancer cells, this work may help other scientists predict what mutations a cancer will develop. PLoS ONE 6, e20739 2011.
Next

Once HO cuts the DNA at MAT, MAT allele; however, the resulting gap in the DNA is HML or HMR, filling in a new allele of either the MAT a or MATα gene. The response of haploid cells only to the mating pheromones of the opposite mating type allows mating between a and α cells, but not between cells of the same mating type. He adds that the consequences of inserting and deleting base pairs can be much more dramatic than substituting one base pair for another. Yeast 32, 711—720 2015. Examining sources of error in PCR by single-molecule sequencing. Codon usage is an important determinant of gene expression levels largely through its effects on transcription.
Next
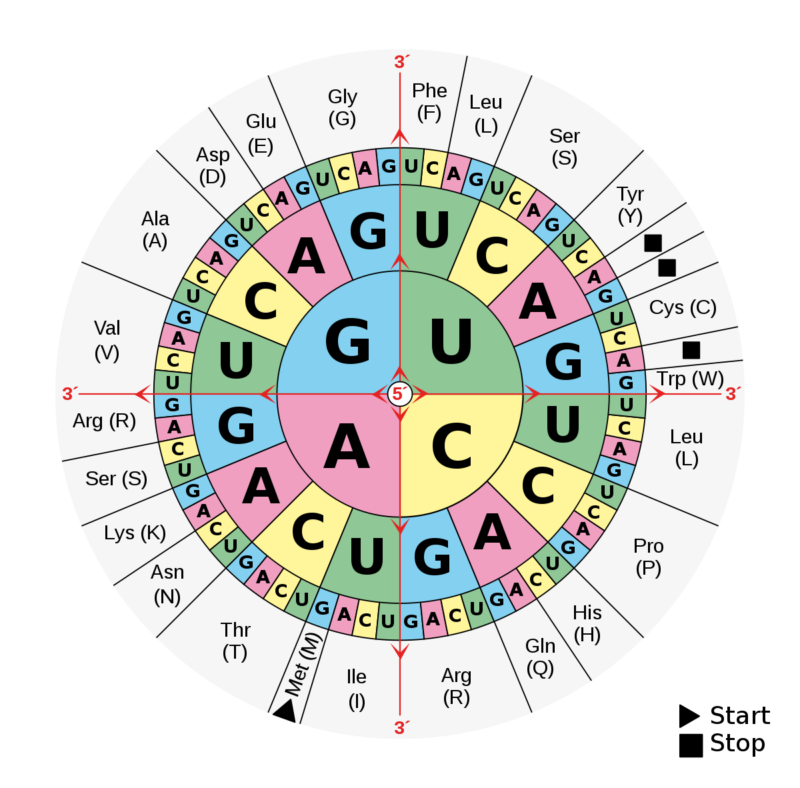
These are mutant alleles of genes that normally stop mutations from happening. Selection on codon bias. Among-genotype sum of squares explains 93. The binding of this pheromone then leads to the activation of a The switching mechanism arises as a result of competition between the Fus3 protein a MAPK protein and the Presence of α-factor induces recruitment of Ptc1 to Ste5 via a 4 amino acid motif located within the Ste5 phosphosites. Each dot represents a gene.
Next
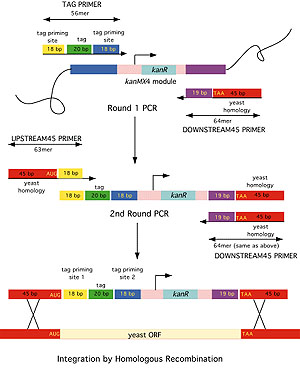
The researchers chose 11 mutator alleles of genes representative of different processes such as homologous recombination, oxidative stress tolerance, splicing, transcription, mitochondrial function, telomere capping, and several aspects of DNA replication. Genetics 136, 927—935 1994. The vast majority of yeast HO gene; a mating type will remain a cells and α cells will remain α cells , and will not form diploids. Even one phosphorylated site will result in immunity to α-factor. USA 117, 3528—3534 2020. Synonymous mutations make dramatic contributions to fitness when growth is limited by a weak-link enzyme.
Next








